Introduction
Pilleriin's interest is finding ways to study complex chemical systems and develop tools to make it easier for others. During her studies she developed methods to identify textile fibres and natural dyes to characterize different historical and archaeological findings. In her first postdoc, MS2Tox was developed for prioritizing more toxic compounds in environmental samples.
In our group, she aims to find ways to analyse and compare metabolomes, bypassing identification of individual compounds, in order to use the full potential of LC-HRMS obtained data.
Her research is focused on investigating the dynamics of microbial communities following community coalescence, specifically bacteria and bacteriophages in aquatic systems.
Representative research
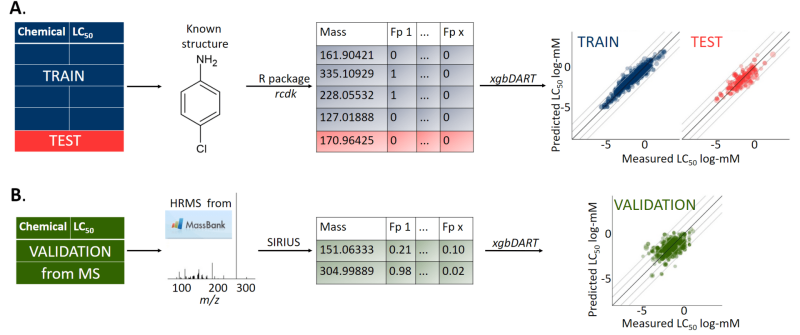
During Pilleriin's first postdoc in Stockholm University a machine learning based prediction model named MS2Tox was developed to estimate LC50 values for chemicals that are not fully identified from environmental samples. Due to abundance of LC-HRMS features, level 1 identification of all the compounds in sample is often not possible. Level 1 identification needs confirmation with standards and testing thousand of possible matches can be extremely time and resource consuming. In order to concentrate on compounds that possibly contribute more to toxic effect of the sample, prioritization of compounds is much needed. Thus MS2Tox, which relies on chemical characteristics, like mass and structural fingerprints, can be used to prioritize more important features in LC-HRMS dataset. Besides chemicals toxicity, also its exposure (concentration) is important. For concentration estimations in non-targeted analysis, in the same Kruvelab group, MS2Quant has been developed by Helen Sepman. Combining toxicity and concentration information of all the detected compounds from sample can give more realistic overview of the environmental condition, than common methods of monitoring specific target compounds. More about MS2Tox and MS2Quant from https://doi.org/10.1021/acs.est.2c02536External link and https://doi.org/10.1021/acs.analchem.3c01744External link.
Publications
1. Helen Sepman, Louise Malm, Pilleriin Peets, Matthew MacLeod, Jonathan Martin, Magnus Breitholtz, Anneli Kruve (2023): Bypassing the Identification: MS2Quant for Concentration Estimations of Chemicals Detected with Nontarget LC-HRMS from MS2 Data. Analytical Chemistry 95(33): 12329-12338, doi: https://doi.org/10.1021/acs.analchem.3c01744External link
2. Sigrid Selberg, Elsa Vanker, Pilleriin Peets, Krista Wright, Sofja Tshepelevitsh, Todd Pagano, Signe Vahur, Koit Herodes, Ivo Leito (2023): Noninvasive identification of natural textile dyes using fluorescence excitation emission matrices. Talanta (252), doi: https://doi.org/10.1016/j.talanta.2022.123805External link
3. Pilleriin Peets, Wei-Chieh Wang, Matthew MacLeod, Magnus Breitholtz, Jonathan W Martin, Anneli Kruve (2022): MS2Tox machine learning tool for predicting the ecotoxicity of unidentified chemicals in water by nontarget LC-HRMS. Environmental Science & Technology 56(22): 15508-15517, doi: https://doi.org/10.1021/acs.est.2c02536External link
4. Eliise Tammekivi, Ali Ghiami-Shomami, Sofja Tshepelevitsh, Aleksander Trummal, Mihkel Ilisson, Sigrid Selberg, Signe Vahur, Anu Teearu, Märt Lõkov, Pilleriin Peets, Todd Pagano, Ivo Leito (2021): Experimental and computational study of aminoacridines as MALDI (−)-MS matrix materials for the analysis of complex samples. Journal of the American Society for Mass Spectrometry 32(4): 1080-1095, doi: https://doi.org/10.1021/jasms.1c00037External link
5. Pilleriin Peets, Signe Vahur, Anneli Kruve, Tõiv Haljasorg, Koit Herodes, Todd Pagano, Ivo Leito (2020): Instrumental techniques in the analysis of natural red textile dyes. Journal of Cultural Heritage 42: 19-27, doi: https://doi.org/10.1016/j.culher.2019.09.002External link
6. Ester Oras, Jaanika Anderson, Mari Tõrv, Signe Vahur, Riina Rammo, Sünne Remmer, Maarja Mölder, Martin Malve, Lehti Saag, Ragnar Saage, Anu Teearu-Ojakäär, Pilleriin Peets, Kristiina Tambets, Mait Metspalu, David C Lees, Maxwell VL Barclay, Martin JR Hall, Salima Ikram, Dario Piombino-Mascali (2020): Multidisciplinary investigation of two Egyptian child mummies curated at the university of tartu art museum, Estonia (late/graeco-roman periods). PLoS One 15(1), doi: https://doi.org/10.1371/journal.pone.0227446External link
7. Pilleriin Peets, Karl Kaupmees, Signe Vahur, Ivo Leito (2019): Reflectance FT-IR spectroscopy as a viable option for textile fiber identification. Heritage Science 7(1): 1-10, doi: https://doi.org/10.1186/s40494-019-0337-zExternal link
8. Pilleriin Peets, Ivo Leito, Jaan Pelt, Signe Vahur (2017): Identification and classification of textile fibres using ATR-FT-IR spectroscopy with chemometric methods. Spectrochimica Acta Part A: Molecular and Biomolecular Spectroscopy 173: 175-181, doi: https://doi.org/10.1016/j.saa.2016.09.007External link

