Dr. Maria Beatriz Walter Costa
Introduction
Bia Walter Costa presenting genome kmers
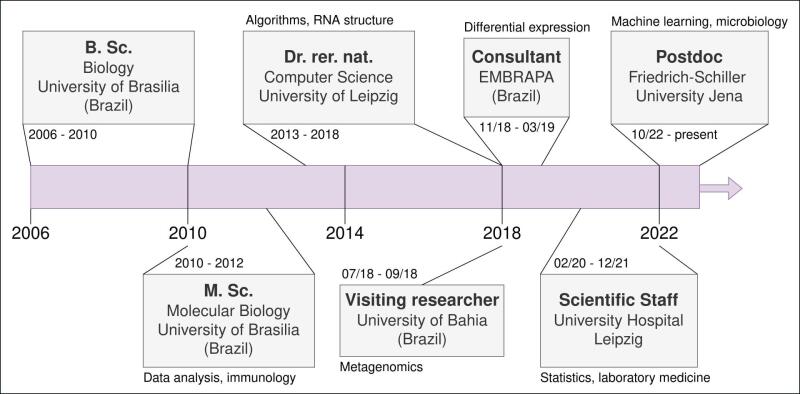
Image: Sybille HuckI am a Bioinformatician and focus on data analysis using Bioinformatics tools such as linux, python and bash. My main interest is to extract valuable information from biological data. For that I collaborate with specialists from different areas of biology.
Currently, I apply machine learning to connect bacterial genomes to their environments. I am using labelled data to build models. With this, I want to better understand how certain genomic features of bacteria allow them to survive in different environments.
Representative research
The SSS-test, Selection on the Secondary Structure test, is a representative research of mine. This test was the first in the scientific community that could detect positive selection in ncRNA structures. I developed it to address an important demand in the field of RNA evolution. Previous research focused on structural conservation, which is used to find functional molecules. However, no tool existed to find positive selection within a conserved group. My tool calculates selection scores based on nucleotide mutations and indels that impact the structure. By applying this tool in ~15 000 long ncRNA orthologs of primates, I found a small set of ~110 human long ncRNAs that have signs of positive selection. This analysis helped colleagues of experimental fields to explore molecules that are important for species-specific traits.
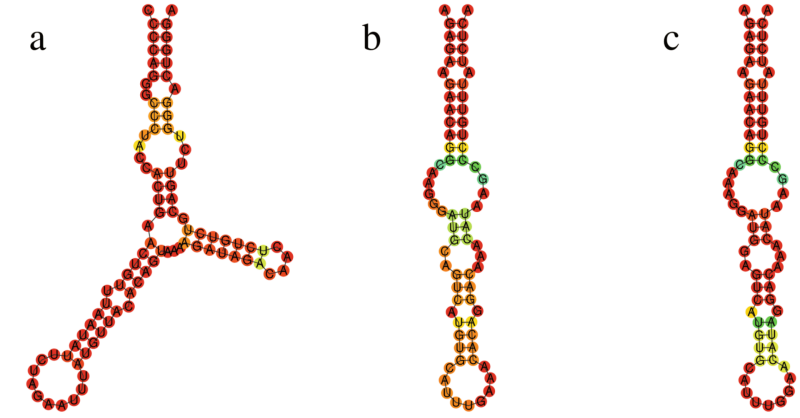
Local structure orthologs of long ncRNA LINC02217. The structure of a) humans has a positive selection signal and is clearly different than the conserved structures of b) chimpanzee/bonobo and c) gorilla. The selection signal was detected by the SSS-test.
Image: Maria Beatriz Walter CostaPublications
- M. B. Walter Costa*, C. Gärtner*, M. Schmidt, T. Berg, D. Seehofer and T. Kaiser (2023): “Revising the MELD Score to Address Sex-Bias in Liver Transplant Prioritization for a German Cohort”External link, Journal of Personalized Medicine 13(6), doi: https://doi.org/10.3390/jpm13060963External link.
- M. B. Walter Costa*, M. Wernsdorfer M*, A. Kehrer A, et al (2021): "The Clinical Decision Support System AMPEL for Laboratory Diagnostics: Implementation and Technical Evaluation"External link, JMIR Medical Informatics 9(6),doi: https://doi.org/10.2196/20407External link.
- M. B. Walter Costa, C. Höner zu Siederdissen, M. Dunjić, P. Stadler, K. Nowick (2019): "SSS-Test: A novel test for detecting selection on the secondary structures of non-coding RNAsExternal link", BMC Bioinformatics 20(151), doi: https://doi.org/10.1186/s12859-019-2711-yExternal link.
- M. B. Walter Costa, C. Höner zu Siederdissen, D Tulpan, P. Stadler, K. Nowick (2018): "Temporal ordering of substitutions in RNA evolution: Uncovering the structural evolution of the Human Accelerated Region 1"External link, Journal of Theoretical Biology 438: 143-150, doi: https://doi.org/10.1016/j.jtbi.2017.11.015External link.
- A. Q. Maranhão, M. B. Costa, L. Guedes, P. M. Moraes-Vieira, T. Raiol, M. M. Brigido (2013): "A Mouse variable gene fragment binds to DNA independently of the BCR context: A Possible role for immature B-Cell repertoire establishmentExternal link", Plos One. doi: https://doi.org/10.1371/journal.pone.0072625External link.
*equal contribution